Lab Data and Databases
Plant Gene Imprinting Database
A collection of imprinting data for a variety of flowering plant species. As much raw data is included as possible. Search by homolog across species.
Single Nuclei Analyses
A GitHub page with all scripts associated with analyzing endosperm single-nuclei data. From our 2021 Nature Plants paper.
Imprinting Analysis
A GitHub page with a suite of scripts we use to analyze and compare genomic imprinting data from reciprocal crosses.
NCBI GEO Series GSE157145
Single nuclei mRNA-seq from Arabidopsis endosperm and seed coat at 4 DAP. Col-Cvi reciprocal crosses. Associated with: Picard CL, Povilus RA, Williams BP, Gehring M. Transcriptional and imprinting complexity in Arabidopsis seeds at single-nucleus resolution. Nat Plants 2021 Jun;7(6):730-738. PMID: 34059805
NCBI GEO Series GSE191307
DNA methylation profiling of wild-type Arabidopsis rosette leaves, cauline leaves, flower buds, and embryos from the same individual plants. DNA methylation profiling and mRNA-seq of leaves of AGL61:DME, dme AGL61:DME, ros1 dml2 dml3 AGL61:DME, and dme ros1 dml2 dml3 AGL61:DME. Associated with: Williams BP, Bechen LL, Pohlmann DA, Gehring M. Somatic DNA demethylation generates tissue-specific methylation states and impacts flowering time. Plant Cell 2022 Mar 29;34(4):1189-1206. PMID: 34954804
NCBI GEO Series GSE197717
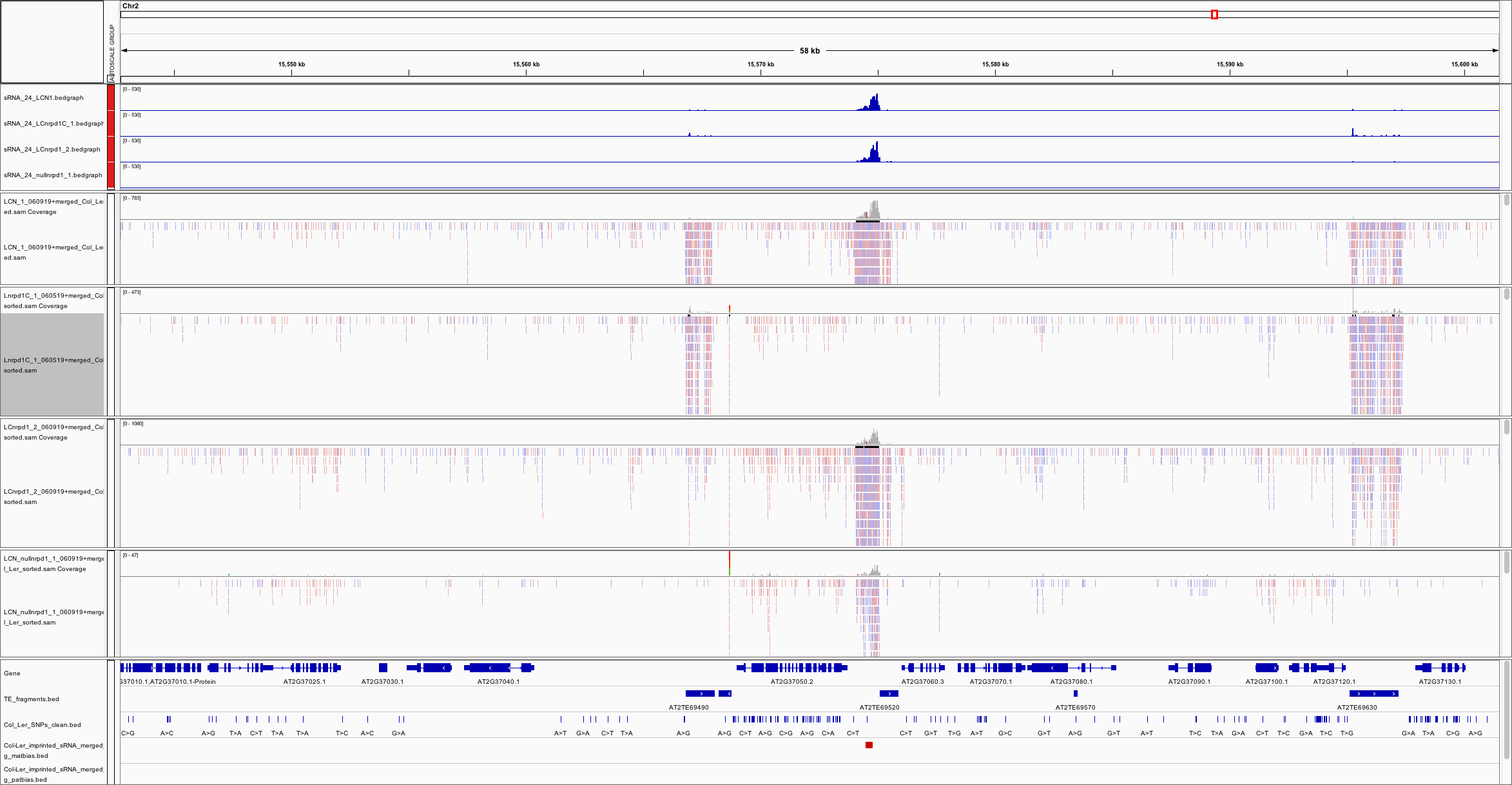
mRNA-seq, small RNA-seq, nanoPARE-seq, and BS-seq profiling of F1 endosperm that inherited an nrpd1 mutation from the mother or the father. Associated with: Satyaki PRV, Gehring M. RNA Pol IV induces antagonistic parent-of-origin effects on Arabidopsis endosperm. PLoS Biol 2022 Apr;20(4):e3001602. PMID: 35389984
NCBI GEO Series GSE126932
mRNA-seq, small RNA-seq, and BS-seq (DNA methylation) profiling of F1 endosperm from diploid x diploid, diploid x tetraploid, diploid x tetraploid nrpd1. Associated with: Satyaki PRV, Gehring M. Paternally Acting Canonical RNA-Directed DNA Methylation Pathway Genes Sensitize Arabidopsis Endosperm to Paternal Genome Dosage. Plant Cell 2019 Jul;31(7):1563-1578. PMID: 31064867
NCBI GEO Series GSE123602
CUT&RUN profiling of H3K27me3, H3, and IgG in Arabidopsis endosperm and leaf. Associated with: Zheng XY, Gehring M. Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reprod 2019 Mar;32(1):63-75. PMID: 30719569
NCBI GEO Series GSE118371
mRNA-Seq in Col-0, Col-0 x Cvi and hdg3-1 Col-0 mutant endosperm. Includes differential expression analysis. Associated with: Pignatta D, Novitzky K, Satyaki PRV, Gehring M. A variably imprinted epiallele impacts seed development. PLoS Genet 2018 Nov;14(11):e1007469. PMID: 30395602
NCBI GEO Series GSE93968
DNA methylation profiling of 10 Col-Cvi RILs, along with parents. mRNA-seq profiling of 2 RILs and parents. Associated with: Picard CL, Gehring M. Proximal methylation features associated with nonrandom changes in gene body methylation. Genome Biol 2017 Apr 26;18(1):73. PMID: 28446217
NCBI GEO Series GSE104240
Leaf whole-genome bisulfite sequencing of plants where the ROS1 epigenetic rheostat as been disrupted, along with controls, across multiple generations. Associated with: Williams BP, Gehring M. Stable transgenerational epigenetic inheritance requires a DNA methylation-sensing circuit. Nat Commun 2017 Dec 14;8(1):2124. PMID: 29242626
NCBI GEO Series GSE94972
small RNA-seq data from endosperm, embryo, and seed coat for Arabidopsis thaliana and Arabidopsis lyrata, including multiple accessions and reciprocal crosses. Also includes mRNA-seq data from WT and nrpd1 mutant endosperm. Associated with: Erdmann RM, Satyaki PRV, Klosinska M, Gehring M. A Small RNA Pathway Mediates Allelic Dosage in Endosperm. Cell Rep 2017 Dec 19;21(12):3364-3372. PMID: 29262317
NCBI GEO Series GSE76076
mRNA-seq from embryo, endosperm, flower buds, and seed coat of Arabidopsis lyrata MN47 and Karhumaki strains and reciprocal crosses. BS-seq data from parental and F1 hybrid endosperm, embryo, and buds. Also includes genome sequencing to identify Karhumaki SNPs. Associated with: Klosinska M, Picard CL, Gehring M. Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat Plants 2016 Sep 19;2:16145. PMID: 27643534
NCBI GEO Series GSE52814
Embryo, endosperm, and whole seed mRNA-seq data from Col, Ler, Cvi reciprocal crosses; whole seed small RNA-seq data from reciprocal crosses; endosperm and embryo whole genome bisulfite sequencing from reciprocal crosses. Used to define imprinted genes and differentially methylated regions. Associated with: Pignatta D, Erdmann RM, Scheer E, Picard CL et al. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife 2014 Jul 3;3:e03198. PMID: 24994762